An example of Bayesian hyperparameter optimization parallelized on cloud VMs using Kubeflow
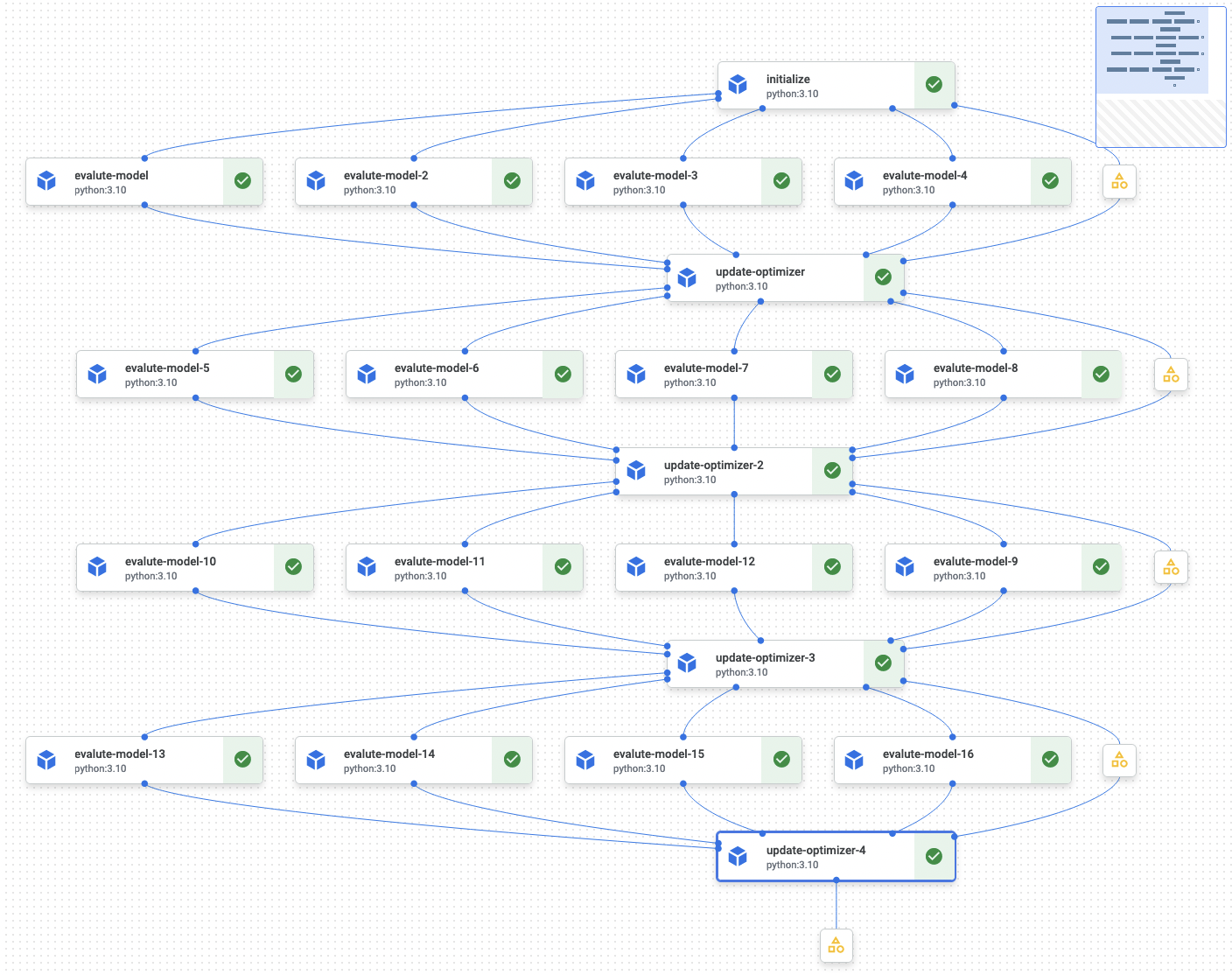
Screenshot of the execution of the Kubeflow pipeline we’ll implement, where each node on the graph corresponds to a cloud virtual machine, and the edges to data that’s passed forward:
To “train” a machine learning model is to carry out some optimization process (e.g. gradient descent). But how do you know which kind of model to optimize in the first place, and which parameters to select for a given model (e.g. number of layers/trees/etc.)? Well, hopefully you can narrow things down a bit via domain knowledge (e.g. “I know that X type of model tends to work well on this type of data”), but unless you are very certain about your guess (or you don’t particularly need to maximise performance), you’ll probably want to carry out some kind of hyperparameter search. Two common techniques for this are grid search and random search, the latter often being used because it would take too long to carry out an exhaustive grid search. A third option is to explicitly treat the hyperparameter search itself as a function, e.g. evaluate_model(hyperparameters) -> score, and to use a “meta” model to optimize this function; we’ll go with this option.
The library Scikit-Optimize has a toy example (reproduced below) of Bayesian optimization that is parallelized using multiple processes on a single machine. However, this single machine approach won’t work well for models that require a lot of resources (e.g. CPU, RAM, GPU), so we’ll adapt the example and parallelize the search across multiple cloud machines (the method could be used to train more serious models on beefier machines, and it wouldn’t take much to adapt it to random search/grid search).
(Why parallelize in the first place?: To be able to run more experiments in a given length of time and so hopefully get better results within that time.)
What is Kubeflow?
Kubeflow is a framework that can be used for writing and running machine learning pipelines. We’ll use its function-based components to do all our work within Python; writing the code the VMs will execute, specifying the data the VMs will pass between eachother, the resources of the VMs, etc. and ultimately compiling all the info into a json and sending it to the cloud.
A nice thing about Kubeflow is it provides the versioning of the execution of code, and the data/artifacts that were part of the execution (as opposed to the versioning of just the code itself, which is git’s domain). This is valuable in the context of machine learning, because a model is a product of not just its code/architecture, but also the data its trained on.
If you happen to have a Kubernetes cluster handy, you could run the pipeline on that, but in this post we’ll use GCP’s managed service, Vertex (services from other cloud providers are available). When we run the pipeline on Vertex, GCP will fire up virtual machines for us, and save artifacts to Cloud Storage buckets. (We pay for what we use.)
Example of Bayesian hyperparameter optimization from Scikit-Optimize docs
Below is the example from the Scikit-Optimize docs that parallelizes the hyperparemeter optimization using multiple CPU cores on a single machine. (Tweaked a bit.)
# We'll use these values in the pipeline as well
NUM_ITERATIONS = 4
NUM_PARALLEL_TRIALS = 4
from skopt import Optimizer, space
from joblib import Parallel, delayed
# "branin" is a function that takes a list of hyperparameter values and returns a score,
# we'll use it in place of a real model.
from skopt.benchmarks import branin
optimizer = Optimizer(
dimensions=[space.Real(-5.0, 10.0), space.Real(0.0, 15.0)],
random_state=1,
base_estimator="gp",
)
all_scores_and_params = []
for i in range(NUM_ITERATIONS):
# Get a list of points in hyperparameter space to evaluate
hyperparam_vals = optimizer.ask(n_points=NUM_PARALLEL_TRIALS)
# Evaluate the points in parallel
scores = Parallel(n_jobs=NUM_PARALLEL_TRIALS)(
delayed(branin)(v) for v in hyperparam_vals
)
all_scores_and_params.extend(zip(hyperparam_vals, scores))
# Update the optimizer with the results
optimizer.tell(hyperparam_vals, scores)
# Print the best score found
print(min(optimizer.yi))
0.8080172110371091
The example from above written with Kubeflow
First import the libraries we’ll use. (kfp is the Kubeflow Pipelines SDK.)
import json
from datetime import datetime
from typing import NamedTuple
from google.cloud import aiplatform
from google.oauth2 import service_account
from kfp.v2 import compiler, dsl
from kfp.v2.dsl import Artifact, Input, Output, pipeline
from pseudo_tuple_component import PseudoTuple, pseudo_tuple_component
Note that pseudo_tuple_component is a Python module I’ve written to
workaround the fact the current version of Kubeflow Pipelines SDK, 1.8,
doesn’t support aggregating the resuls of multiple components.
It involves use of the Python’s inspect, and ast, modules
to modify the source code of a function… Code here.
PIPELINE_NAME = "scikit-opt-example-pipeline"
with open("vertex_config.json", "r") as f:
gcp_cfg = json.load(f) # I put GCP related stuff in here
credentials = service_account.Credentials.from_service_account_file(
gcp_cfg["credentials_path"]
)
Below we’ll define “components”, which are the things that run on a single cloud VM. Note that the imports need to go inside the function, because ultimately the contents of the function will be dumped into a string by kfp and run from within a Docker container. Also the type hints are significant, because kfp uses them to work out how to deal with the inputs and outputs to the VMs (and there’s limitations to what can be used).
@dsl.component(
packages_to_install=["scikit-optimize==0.9.0", "dill==0.3.6"],
base_image="python:3.10",
)
def initialize(
random_state: int,
n_points: int,
optimizer_out: Output[Artifact],
) -> NamedTuple("Outputs", [("hyperparam_vals", str)]):
"""Initialize the optimizer and get the first set of hyperparameter values to evaluate."""
import json
import dill
from skopt import Optimizer, space
optimizer = Optimizer(
dimensions=[space.Real(-5.0, 10.0), space.Real(0.0, 15.0)],
random_state=random_state,
base_estimator="gp",
)
hyperparam_vals = optimizer.ask(n_points=n_points)
with open(optimizer_out.path, "wb") as f:
dill.dump(optimizer, f)
return (json.dumps(hyperparam_vals),)
@dsl.component(
packages_to_install=["scikit-optimize==0.9.0", "dill==0.3.6"],
base_image="python:3.10",
)
def evalute_model(
hyperparam_vals: str,
idx: int,
) -> float:
"""Evaluate a model with the given hyperparameter values."""
import json
from skopt.benchmarks import branin
params = json.loads(hyperparam_vals)[idx]
score = float(branin(params))
return score
# `pseudo_tuple_component` is a custom component I wrote to work around
# the fact that KFP doesn't support tuples
# of kubeflow artifacts as function args.
@pseudo_tuple_component(
packages_to_install=["scikit-optimize==0.9.0", "dill==0.3.6"],
base_image="python:3.10",
globals_=globals(),
locals_=locals(),
)
def update_optimizer(
optimizer_in: Input[Artifact],
hyperparam_vals: str,
scores: PseudoTuple(NUM_PARALLEL_TRIALS, float),
optimizer_out: Output[Artifact],
) -> NamedTuple("Outputs", [("hyperparam_vals", str), ("best_score_found", float)]):
"""Update the optimizer with the results of the previous evaluation
and get the next set of hyperparameter values to evaluate."""
import json
import dill
with open(optimizer_in.path, "rb") as f:
optimizer = dill.load(f)
optimizer.tell(json.loads(hyperparam_vals), scores)
hyperparam_vals = optimizer.ask(n_points=4)
with open(optimizer_out.path, "wb") as f:
dill.dump(optimizer, f)
return json.dumps(hyperparam_vals), min(optimizer.yi)
Next we’ll define the pipeline. Note it isn’t actually executed until we compile it and send it to the cloud. We’re basically specifying which components to run in what order, and what to pass to each component. We also specify resources for the VMs that will run the components here, (but it’s up to the cloud provider whether they respect it, e.g. GCP hasn’t given me a machine smaller than 2 CPU, 16GB ram).
@pipeline(
name=PIPELINE_NAME,
pipeline_root=gcp_cfg["pipeline_root"],
)
def my_pipeline(random_state: int = 1):
initialize_ = (
initialize(random_state=random_state, n_points=NUM_PARALLEL_TRIALS)
.set_memory_limit("8G")
.set_cpu_limit("1")
)
latest_optimizer = initialize_
for i in range(NUM_ITERATIONS):
scores = {}
for i in range(NUM_PARALLEL_TRIALS):
evalute_model_ = (
evalute_model(
hyperparam_vals=latest_optimizer.outputs["hyperparam_vals"], idx=i
)
.set_memory_limit("8G")
.set_cpu_limit("1")
)
scores[f"scores_{i}"] = evalute_model_.output
latest_optimizer = (
update_optimizer(
optimizer_in=latest_optimizer.outputs["optimizer_out"],
hyperparam_vals=latest_optimizer.outputs["hyperparam_vals"],
**scores,
)
.set_memory_limit("8G")
.set_cpu_limit("1")
)
# compile the pipeline into a json that contains
# everything needed to run the pipeline
compiler.Compiler().compile(
pipeline_func=my_pipeline, package_path=f"{PIPELINE_NAME}.json"
)
Now it’s a matter of sending the pipeline json to the cloud:
TIMESTAMP = datetime.now().strftime("%Y%m%d%H%M%S")
job = aiplatform.PipelineJob(
display_name=f"{PIPELINE_NAME}_job",
credentials=credentials,
template_path=f"{PIPELINE_NAME}.json",
job_id=f"{PIPELINE_NAME}-{TIMESTAMP}",
pipeline_root=gcp_cfg["pipeline_root"],
enable_caching=True,
project=gcp_cfg["project_id"],
location=gcp_cfg["region"],
)
job.submit(
service_account=gcp_cfg["service_account"], experiment=gcp_cfg["experiment_name"]
)
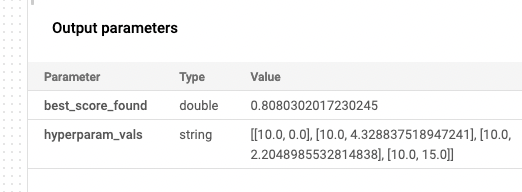
And… after some time… we get the our result, 0.8080302017230245, which is close enough to our local result of 0.8080172110371091.
References:
(In no particular order)
- https://scikit-optimize.github.io/0.9/auto_examples/parallel-optimization.html
- https://codelabs.developers.google.com/vertex-pipelines-intro
- https://www.cloudskillsboost.google/focuses/21234?parent=catalog
- https://www.kubeflow.org/docs/components/pipelines/v1/sdk-v2/python-function-components/
- https://www.kubeflow.org/docs/components/pipelines/v2/author-a-pipeline/component-io/
- https://stackoverflow.com/questions/70358400/kubeflow-vs-vertex-ai-pipelines
- “KubeFlow pipeline stages take a lot less to set up than Vertex in my experience (seconds vs couple of minutes). This was expected, as stages are just containers in KF, and it seems in Vertex full-fledged instances are provisioned to run the containers”
Regarding the necessity of writing pseudo_tuple_component.. Kubeflow has dsl.ParallelFor,
but there doesn’t seem to be a way to aggregate results (see Kubeflow issues 1933, 3412; and this stackoverflow uses kubeflow v1, but vertex requires kubeflow v2 and where this doesn’t work).